Reading and writing fields#
There are three file formats to which a discretisedfield.Field object can be saved:
OOMMF Vector Field File Format (OVF) for exchanging fields with micromagnetic simulators.
HDF5
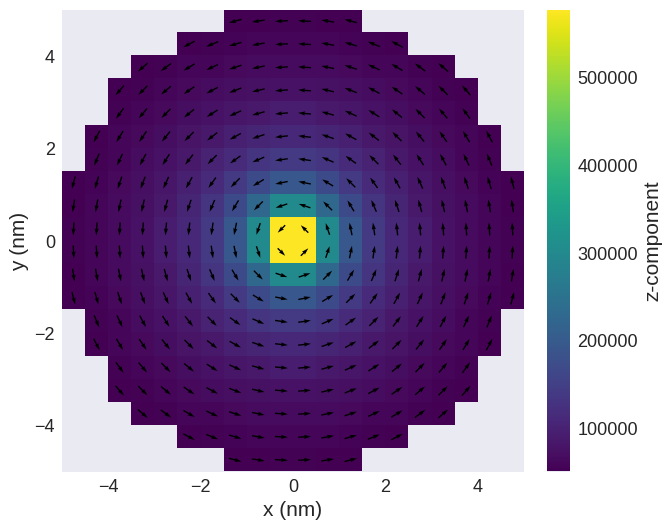
Let us say we have a nanosphere sample:
with \(r=5\,\text{nm}\). The space is discretised into cells with dimensions \((0.5\,\text{nm}, 0.5\,\text{nm}, 0.5\,\text{nm})\). The value of the field at \((x, y, z)\) point is \((-cy, cx, cz)\), with \(c=10^{9}\). The norm of the field inside the cylinder is \(10^{6}\).
Let us first build that field.
[1]:
import discretisedfield as df
r = 5e-9
cell = (0.5e-9, 0.5e-9, 0.5e-9)
mesh = df.Mesh(p1=(-r, -r, -r), p2=(r, r, r), cell=cell)
def norm_fun(pos):
x, y, z = pos
if x**2 + y**2 + z**2 <= r**2:
return 1e6
else:
return 0
def value_fun(pos):
x, y, z = pos
c = 1e9
return (-c * y, c * x, c * z)
field = df.Field(mesh, nvdim=3, value=value_fun, norm=norm_fun, valid="norm")
field.sel("z").mpl()
Writing the field to a file#
The main method used for saving fields in different files is discretisedfield.Field.to_file(). It takes filename as an argument, which is a string with one of the following extensions:
'.vtk'for saving in the VTK format'.ovf','.omf','.ohf'for saving in the OVF format'.h5'or'.hdf5'for saving and HDF5 file
Let us firstly save the field in the VTK file format. There are three different possible representations: bin, txt and xml. Refer to the VTK documentation for details. By default, VTK files in discretisedfield are saved as bin.
[2]:
vtkfilename = "my_vtk_file.vtk"
field.to_file(vtkfilename)
We can check if the file was saved in the current directory.
[3]:
import os
os.path.isfile(f"./{vtkfilename}")
[3]:
True
Now, we can delete the file:
[4]:
os.remove(f"./{vtkfilename}")
Next, we can save the field in the OVF format and check whether it was created in the current directory.
[5]:
omffilename = "my_omf_file.omf"
field.to_file(omffilename)
os.path.isfile(f"./{omffilename}")
[5]:
True
There are three different possible representations of an OVF file: one ASCII (txt) and two binary (bin4 or bin8). Binary bin8 representation is the default representation when discretisedfield.Field.to_file() is called. If a different representation is required, it can be passed via representation argument.
[6]:
field.to_file(omffilename, representation="bin4")
os.path.isfile(f"./{omffilename}")
[6]:
True
In the same way, we can save the HDF5 file:
[7]:
hdf5filename = "my_omf_file.hdf5"
field.to_file(hdf5filename)
os.path.isfile(f"./{hdf5filename}")
[7]:
True
Reading files#
The method for reading OVF files is a class method discretisedfield.Field.from_file(). By passing a filename argument, it reads the file and creates a discretisedfield.Field object. It is not required to pass the representation of the OVF and the VTK file to the discretisedfield.Field.from_file() method, because it can retrieve it from the content of the file.
[8]:
read_field = df.Field.from_file(omffilename)
Like previouly, we can quickly visualise the field
[9]:
read_field.sel("z").mpl()
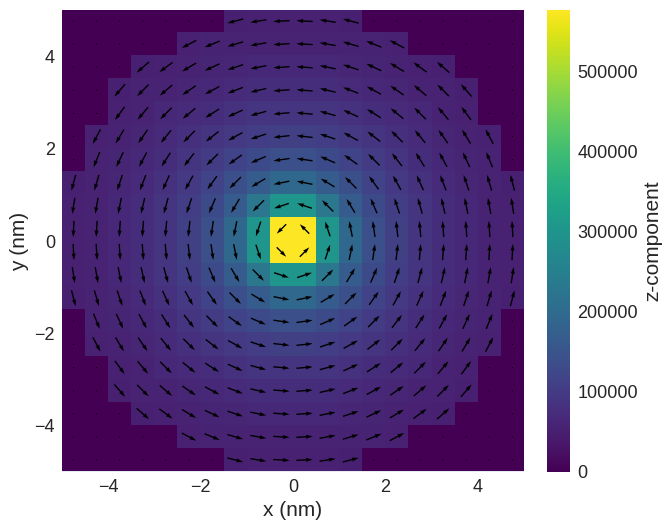
We can see that the OVF (and VTK) file cannot store the valid array and hence this data is lost.
In HDF5 files, we have more freedom and can also store the valid information.
[10]:
read_file = df.Field.from_file(hdf5filename)
read_file.sel("z").mpl()
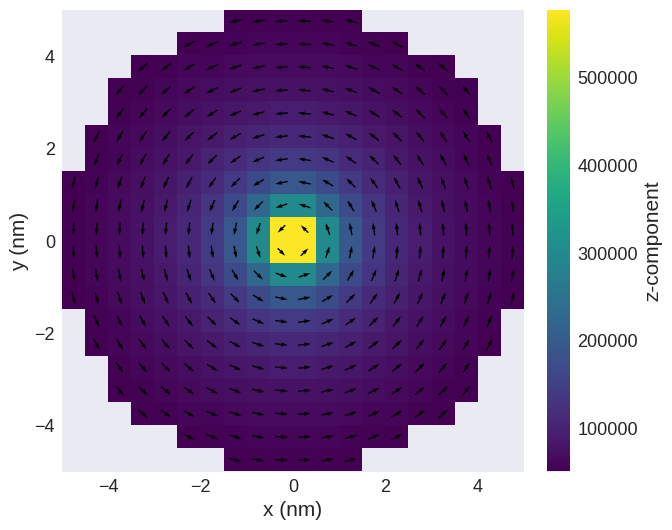
Finally, we can delete the OVF and HDF5 file we created.
[11]:
os.remove(f"./{omffilename}")
[12]:
os.remove(f"./{hdf5filename}")
Subregions#
If we define subregions on the mesh, they are stored as follows:
- For OVF and VTK we save an additional json file that contains the subregion information. If we save the field to
our_field.vtkthe subregion information is stored asour_field.vtk.subregions.json. When reading a file,discretisedfieldautomatically checks if there is an additional subregion json file and loads the additional data from there.Note: Be careful to remove old subregion json files if you overwrite an existing file on disk where the field had subregions with a new field that does not have subregions. Otherwise, reading will potentially fail because the old subregions are incompatible with the new mesh. The HDF5 file directly contains all subregion information and no additional file is required. For more details about its format, please refer to our HDF5 specification.


