Analysing simulation results#
With micromagneticdata we can analyse simulation results created with oommfc. This notebook summarises the available functionality.
[1]:
import os
import micromagneticdata as md
We have a set of example simulations stored in the test directory of micromagneticdata that we use to demonstrate its functionality.
[2]:
dirname = os.path.join("..", "micromagneticdata", "tests", "test_sample")
Data#
First, we creata a Data object. We need to pass the name of the micromagneticmodel.System that we used to run the simulation and optionally an additional path to the base directory.
[3]:
data = md.Data(name="rectangle", dirname=dirname)
The Data object contains all simulation runs of the System. These are called drives.
[4]:
data.n
[4]:
7
[5]:
data.info
[5]:
| drive_number | date | time | start_time | adapter | adapter_version | driver | t | n | end_time | elapsed_time | success | info.json | output_step | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 2025-05-31 | 22:02:42 | 2025-05-31T22:02:42 | oommfc | 0.65.0 | TimeDriver | 0.0 | 25 | 2025-05-31T22:02:42 | 00:00:01 | True | available | <NA> |
| 1 | 1 | 2025-05-31 | 22:02:42 | 2025-05-31T22:02:42 | mumax3c | 0.3.0 | TimeDriver | 0.0 | 15 | 2025-05-31T22:02:42 | 00:00:01 | True | available | <NA> |
| 2 | 2 | 2025-05-31 | 22:02:42 | 2025-05-31T22:02:42 | mumax3c | 0.3.0 | TimeDriver | 0.0 | 250 | 2025-05-31T22:02:43 | 00:00:01 | True | available | <NA> |
| 3 | 3 | 2025-05-31 | 22:02:43 | 2025-05-31T22:02:43 | mumax3c | 0.3.0 | RelaxDriver | <NA> | <NA> | 2025-05-31T22:02:43 | 00:00:01 | True | available | <NA> |
| 4 | 4 | 2025-05-31 | 22:02:43 | 2025-05-31T22:02:43 | mumax3c | 0.3.0 | MinDriver | <NA> | <NA> | 2025-05-31T22:02:43 | 00:00:01 | True | available | <NA> |
| 5 | 5 | 2025-05-31 | 22:02:43 | 2025-05-31T22:02:43 | oommfc | 0.65.0 | TimeDriver | 0.0 | 5 | 2025-05-31T22:02:44 | 00:00:01 | True | available | <NA> |
| 6 | 6 | 2025-05-31 | 22:02:44 | 2025-05-31T22:02:44 | oommfc | 0.65.0 | MinDriver | <NA> | <NA> | 2025-05-31T22:02:44 | 00:00:01 | True | available | True |
Drive#
To access one Drive we can index the Data object.
[6]:
drive = data[6]
The Drive object has a number of properties containing information about the drive.
[7]:
drive.n
[7]:
14
[8]:
drive.info
[8]:
{'drive_number': 6,
'date': '2025-05-31',
'time': '22:02:44',
'start_time': '2025-05-31T22:02:44',
'adapter': 'oommfc',
'adapter_version': '0.65.0',
'driver': 'MinDriver',
'output_step': True,
'end_time': '2025-05-31T22:02:44',
'elapsed_time': '00:00:01',
'success': True}
[9]:
drive = data[6]
[10]:
drive.x
[10]:
'iteration'
[11]:
drive.info
[11]:
{'drive_number': 6,
'date': '2025-05-31',
'time': '22:02:44',
'start_time': '2025-05-31T22:02:44',
'adapter': 'oommfc',
'adapter_version': '0.65.0',
'driver': 'MinDriver',
'output_step': True,
'end_time': '2025-05-31T22:02:44',
'elapsed_time': '00:00:01',
'success': True}
[12]:
print(drive.calculator_script)
# MIF 2.2
SetOptions {
basename rectangle
scalar_output_format %.12g
scalar_field_output_format {binary 8}
vector_field_output_format {binary 8}
}
# BoxAtlas for total_atlas
Specify Oxs_BoxAtlas:total_atlas {
xrange { -5e-08 5e-08 }
yrange { -2.5e-08 2.5e-08 }
zrange { 0.0 2e-08 }
name total
}
# BoxAtlas for entire_atlas
Specify Oxs_BoxAtlas:entire_atlas {
xrange { -5e-08 5e-08 }
yrange { -2.5e-08 2.5e-08 }
zrange { 0.0 2e-08 }
name entire
}
# MultiAtlas
Specify Oxs_MultiAtlas:main_atlas {
atlas :total_atlas
atlas :entire_atlas
xrange { -5e-08 5e-08 }
yrange { -2.5e-08 2.5e-08 }
zrange { 0.0 2e-08 }
}
# RectangularMesh
Specify Oxs_RectangularMesh:mesh {
cellsize { 5e-09 5e-09 5e-09 }
atlas :main_atlas
}
# UniformExchange
Specify Oxs_UniformExchange:exchange {
A 1.3e-11
}
# FixedZeeman
Specify Oxs_FixedZeeman:zeeman {
field {0.0 0.0 1000000.0}
}
# m0 file
Specify Oxs_FileVectorField:m0 {
file m0.omf
atlas :main_atlas
}
# m0_norm
Specify Oxs_VecMagScalarField:m0_norm {
field :m0
}
# CGEvolver
Specify Oxs_CGEvolve:evolver {
}
# MinDriver
Specify Oxs_MinDriver {
evolver :evolver
mesh :mesh
Ms :m0_norm
m0 :m0
stopping_mxHxm 0.1
}
Destination table mmArchive
Destination mags mmArchive
Schedule DataTable table Step 1
Schedule Oxs_MinDriver::Magnetization mags Step 1
The initial magnetisation can be obtained with m0. It returns a discretisedfield.Field object.
[13]:
drive.m0
[13]:
- Mesh
- Region
- pmin = [-5e-08, -2.5e-08, 0.0]
- pmax = [5e-08, 2.5e-08, 2e-08]
- dims = ['x', 'y', 'z']
- units = ['m', 'm', 'm']
- n = [20, 10, 4]
- subregions:
- Region total
- pmin = [-5e-08, -2.5e-08, 0.0]
- pmax = [5e-08, 2.5e-08, 2e-08]
- dims = ['x', 'y', 'z']
- units = ['m', 'm', 'm']
- Region total
- Region
- nvdim = 3
- vdims:
- x
- y
- z
- unit = None
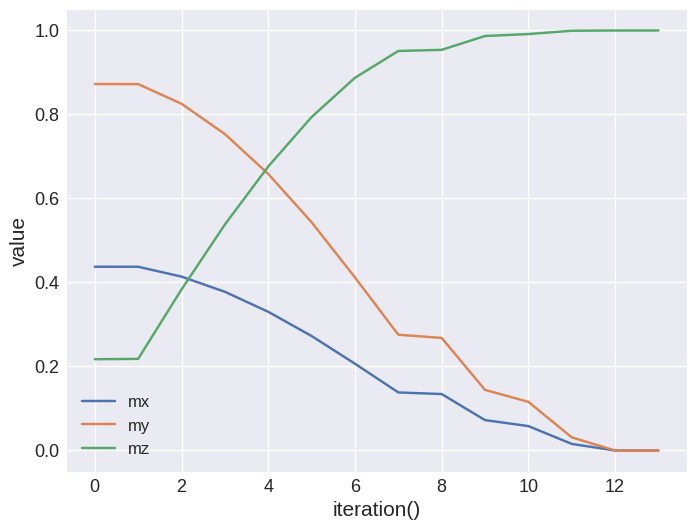
Scalar data of the drive can be accessed via the table property that returns a ubermagtable.Table object.
[14]:
drive.table.mpl(y=["mx", "my", "mz"])
[15]:
drive.n
[15]:
14
We can access the magnetisation at a single step or iterate over the drive.
[16]:
drive[0]
[16]:
- Mesh
- Region
- pmin = [-5e-08, -2.5e-08, 0.0]
- pmax = [5e-08, 2.5e-08, 2e-08]
- dims = ['x', 'y', 'z']
- units = ['m', 'm', 'm']
- n = [20, 10, 4]
- subregions:
- Region total
- pmin = [-5e-08, -2.5e-08, 0.0]
- pmax = [5e-08, 2.5e-08, 2e-08]
- dims = ['x', 'y', 'z']
- units = ['m', 'm', 'm']
- Region total
- Region
- nvdim = 3
- vdims:
- x
- y
- z
- unit = A/m
[17]:
list(drive)
[17]:
[Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m),
Field(Mesh(Region(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']), n=[20, 10, 4], subregions: (Region`total`(pmin=[-5e-08, -2.5e-08, 0.0], pmax=[5e-08, 2.5e-08, 2e-08], dims=['x', 'y', 'z'], units=['m', 'm', 'm']))), nvdim=3, vdims: (x, y, z), unit=A/m)]
We can also select a parts of the data and get a new drive, e.g. every second of the first 10 elements.
[18]:
drive_selection = drive[:10:2]
drive_selection
[18]:
OOMMFDrive(name='rectangle', number=6, dirname='../micromagneticdata/tests/test_sample', x='iteration')
[19]:
drive_selection.n
[19]:
5
It is possible to convert all magnetisation files into vtk for visualisation e.g. in Paraview.
[20]:
# drive.ovf2vtk()
We can convert a drive into an xarray.DataArray. An additional dimension is added depending on the type of drive.
[21]:
drive.to_xarray()
[21]:
<xarray.DataArray 'field' (iteration: 14, x: 20, y: 10, z: 4, vdims: 3)>
array([[[[[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05],
[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05],
[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05],
[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05]],
[[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05],
[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05],
[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05],
[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05]],
[[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05],
[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05],
[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05],
[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05]],
...,
[[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05],
[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05],
[ 3.50067876e+05, 6.98022376e+05, 1.73831082e+05],
...
[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05],
[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05],
[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05]],
...,
[[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05],
[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05],
[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05],
[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05]],
[[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05],
[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05],
[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05],
[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05]],
[[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05],
[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05],
[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05],
[-3.41568167e-06, -6.81074074e-06, 8.00000000e+05]]]]])
Coordinates:
* x (x) float64 -4.75e-08 -4.25e-08 -3.75e-08 ... 4.25e-08 4.75e-08
* y (y) float64 -2.25e-08 -1.75e-08 -1.25e-08 ... 1.75e-08 2.25e-08
* z (z) float64 2.5e-09 7.5e-09 1.25e-08 1.75e-08
* vdims (vdims) <U1 'x' 'y' 'z'
* iteration (iteration) float64 0.0 1.0 2.0 3.0 4.0 ... 10.0 11.0 12.0 13.0
Attributes:
drive_number: 6
date: 2025-05-31
time: 22:02:44
start_time: 2025-05-31T22:02:44
adapter: oommfc
adapter_version: 0.65.0
driver: MinDriver
output_step: True
end_time: 2025-05-31T22:02:44
elapsed_time: 00:00:01
success: TrueCombinedDrive#
Multiple drives can be concatenated to create one “longer” drive. This can e.g. be convenient to analyse multiple consecutive time drives. Combining drives is done via the << operator which appends the right-hand-side drive to the left-hand-side drive. It returns a new CombinedDrive object that behaves very similar to the Drive object.
[22]:
combined = data[0] << data[1] << data[2]
combined
[22]:
CombinedDrive(
OOMMFDrive(name='rectangle', number=0, dirname='../micromagneticdata/tests/test_sample', x='t'),
Mumax3Drive(name='rectangle', number=1, dirname='../micromagneticdata/tests/test_sample', x='t'),
Mumax3Drive(name='rectangle', number=2, dirname='../micromagneticdata/tests/test_sample', x='t')
)
The combined drive has a property drives that provides a list of all individual drives.
[23]:
combined.drives
[23]:
(OOMMFDrive(name='rectangle', number=0, dirname='../micromagneticdata/tests/test_sample', x='t'),
Mumax3Drive(name='rectangle', number=1, dirname='../micromagneticdata/tests/test_sample', x='t'),
Mumax3Drive(name='rectangle', number=2, dirname='../micromagneticdata/tests/test_sample', x='t'))
[24]:
combined.info
[24]:
{'drive_numbers': [0, 1, 2], 'driver': 'TimeDriver'}
We have direct access to the initial magnetisation of the first drive.
[25]:
combined.m0
[25]:
- Mesh
- Region
- pmin = [-5e-08, -2.5e-08, 0.0]
- pmax = [5e-08, 2.5e-08, 2e-08]
- dims = ['x', 'y', 'z']
- units = ['m', 'm', 'm']
- n = [20, 10, 4]
- subregions:
- Region total
- pmin = [-5e-08, -2.5e-08, 0.0]
- pmax = [5e-08, 2.5e-08, 2e-08]
- dims = ['x', 'y', 'z']
- units = ['m', 'm', 'm']
- Region total
- Region
- nvdim = 3
- vdims:
- x
- y
- z
- unit = None
[26]:
combined.n
[26]:
290
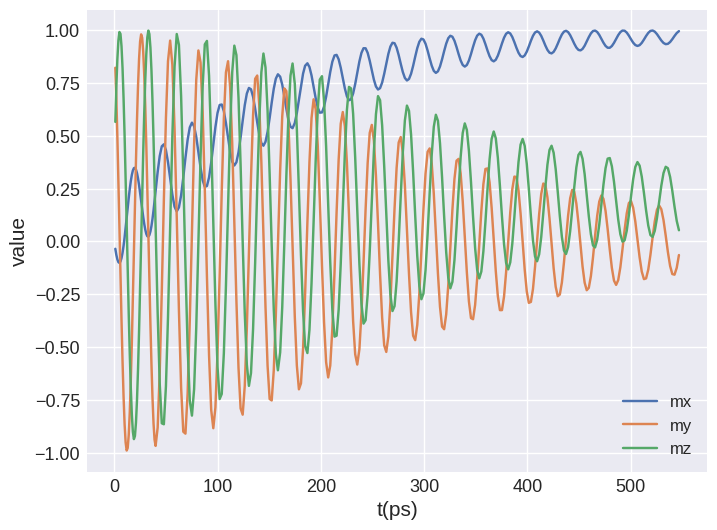
The combined drive has one large table that contains the data for all individual drives.
[27]:
combined.table.mpl(y=["mx", "my", "mz"])
We can iterate over the drives in the combined drive or access a single element.
[28]:
combined[15]
[28]:
- Mesh
- Region
- pmin = [-5e-08, -2.5e-08, 0.0]
- pmax = [5e-08, 2.5e-08, 2e-08]
- dims = ['x', 'y', 'z']
- units = ['m', 'm', 'm']
- n = [20, 10, 4]
- subregions:
- Region total
- pmin = [-5e-08, -2.5e-08, 0.0]
- pmax = [5e-08, 2.5e-08, 2e-08]
- dims = ['x', 'y', 'z']
- units = ['m', 'm', 'm']
- Region total
- Region
- nvdim = 3
- vdims:
- x
- y
- z
- unit = A/m
[29]:
combined.x
[29]:
't'
The combined drive can be converted into an xarray.DataArray similar to the normal Drive.
[30]:
combined.to_xarray()
[30]:
<xarray.DataArray 'field' (t: 290, x: 20, y: 10, z: 4, vdims: 3)>
array([[[[[ -27584.49540021, 658579.77501927, 453334.06616874],
[ -27584.49540021, 658579.77501927, 453334.06616874],
[ -27584.49540021, 658579.77501927, 453334.06616874],
[ -27584.49540021, 658579.77501927, 453334.06616874]],
[[ -27584.49540021, 658579.77501927, 453334.06616874],
[ -27584.49540021, 658579.77501927, 453334.06616874],
[ -27584.49540021, 658579.77501927, 453334.06616874],
[ -27584.49540021, 658579.77501927, 453334.06616874]],
[[ -27584.49540021, 658579.77501927, 453334.06616874],
[ -27584.49540021, 658579.77501927, 453334.06616874],
[ -27584.49540021, 658579.77501927, 453334.06616874],
[ -27584.49540021, 658579.77501927, 453334.06616874]],
...,
[[ -27584.49540021, 658579.77501927, 453334.06616874],
[ -27584.49540021, 658579.77501927, 453334.06616874],
[ -27584.49540021, 658579.77501927, 453334.06616874],
...
[ 797212.875 , -51317.65625 , 42639.36328125],
[ 797212.875 , -51317.65625 , 42639.36328125],
[ 797212.875 , -51317.65625 , 42639.36328125]],
...,
[[ 797212.875 , -51317.65625 , 42639.36328125],
[ 797212.875 , -51317.65625 , 42639.36328125],
[ 797212.875 , -51317.65625 , 42639.36328125],
[ 797212.875 , -51317.65625 , 42639.36328125]],
[[ 797212.875 , -51317.65625 , 42639.36328125],
[ 797212.875 , -51317.65625 , 42639.36328125],
[ 797212.875 , -51317.65625 , 42639.36328125],
[ 797212.875 , -51317.65625 , 42639.36328125]],
[[ 797212.875 , -51317.65625 , 42639.36328125],
[ 797212.875 , -51317.65625 , 42639.36328125],
[ 797212.875 , -51317.65625 , 42639.36328125],
[ 797212.875 , -51317.65625 , 42639.36328125]]]]])
Coordinates:
* x (x) float64 -4.75e-08 -4.25e-08 -3.75e-08 ... 4.25e-08 4.75e-08
* y (y) float64 -2.25e-08 -1.75e-08 -1.25e-08 ... 1.75e-08 2.25e-08
* z (z) float64 2.5e-09 7.5e-09 1.25e-08 1.75e-08
* vdims (vdims) <U1 'x' 'y' 'z'
* t (t) float64 1e-12 2e-12 3e-12 ... 5.423e-10 5.445e-10 5.466e-10
Attributes:
drive_numbers: [0, 1, 2]
driver: TimeDriverWe can also append an other drive to a combined drive.
[31]:
combined << data[0]
[31]:
CombinedDrive(
OOMMFDrive(name='rectangle', number=0, dirname='../micromagneticdata/tests/test_sample', x='t'),
Mumax3Drive(name='rectangle', number=1, dirname='../micromagneticdata/tests/test_sample', x='t'),
Mumax3Drive(name='rectangle', number=2, dirname='../micromagneticdata/tests/test_sample', x='t'),
OOMMFDrive(name='rectangle', number=0, dirname='../micromagneticdata/tests/test_sample', x='t')
)
Widgets#
[32]:
data.selector()
[32]:
[33]:
drive.slider()
[33]:


