Multiple energy terms of the same class#
Here we demonstrate how to define multiple energy terms of the same class in the energy equation. For the sample, we choose a one-dimensional chain of magnetic moments.
[1]:
import random
import oommfc as mc
import discretisedfield as df
import micromagneticmodel as mm
p1 = (-10e-9, 0, 0)
p2 = (10e-9, 1e-9, 1e-9)
cell = (1e-9, 1e-9, 1e-9)
region = df.Region(p1=p1, p2=p2)
mesh = df.Mesh(region=region, cell=cell)
The mesh is
[2]:
mesh.mpl(box_aspect=(10, 4, 4), figsize=(8, 8))
Let us say that the system has an energy equation, which consists of two Zeeman energy terms.
[3]:
H1 = (0, 0, 1e6)
H2 = (1e6, 0, 0)
system = mm.System(name="multiple_terms")
Now, if we try to add two Zeeman energy terms, we get an exception raised.
[4]:
system.energy = mm.Zeeman(H=(0, 0, 1e5)) + mm.Zeeman(H=(0, 1e5, 0))
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[4], line 1
----> 1 system.energy = mm.Zeeman(H=(0, 0, 1e5)) + mm.Zeeman(H=(0, 1e5, 0))
File ~/repos/ubermag-devtools/repos/micromagneticmodel/micromagneticmodel/abstract/term.py:115, in Term.__add__(self, other)
113 result = getattr(mm, self._container_class)()
114 result += self
--> 115 result += other
116 return result
File ~/repos/ubermag-devtools/repos/micromagneticmodel/micromagneticmodel/abstract/container.py:378, in Container.__add__(self, other)
369 if any(
370 isinstance(other, term.__class__) and other.name == term.name
371 for term in self._terms
372 ):
373 msg = (
374 f"There is already a term of type {other.__class__} with name "
375 f"'{other.name}' in {self.__class__}. Please provide a different "
376 f"name for {other}."
377 )
--> 378 raise ValueError(msg)
379 else:
380 result._terms.append(other)
ValueError: There is already a term of type <class 'micromagneticmodel.energy.zeeman.Zeeman'> with name 'zeeman' in <class 'micromagneticmodel.energy.energy.Energy'>. Please provide a different name for Zeeman(H=(0, 100000.0, 0)).
This is because different energy terms must have different names, so they can be uniquely identified. So, we have to give names to our energy terms:
[5]:
system.energy = mm.Zeeman(H=H1, name="zeeman1") + mm.Zeeman(H=H2, name="zeeman2")
We need to define the system’s magnetisation (system.m). We are going to make it random with \(M_\text{s}=8\times10^{5} \,\text{Am}^{-1}\)
[6]:
random.seed(1)
Ms = 8e5 # saturation magnetisation (A/m)
def m_fun(pos):
return [2 * random.random() - 1 for i in range(3)]
system.m = df.Field(mesh, nvdim=3, value=m_fun, norm=Ms)
Now, we can minimise the system’s energy:
[7]:
md = mc.MinDriver()
md.drive(system)
Running OOMMF (ExeOOMMFRunner)[2023/10/23 16:08]... (0.4 s)
We expect that now all magnetic moments are aligned parallel or antiparallel to the anisotropy axis (in the \(z\)-direction).
[8]:
system.m.sel("y").mpl(
figsize=(10, 3), scalar_kw={"clim": [-0.1, 0.1], "colorbar_label": "$m_y$"}
)
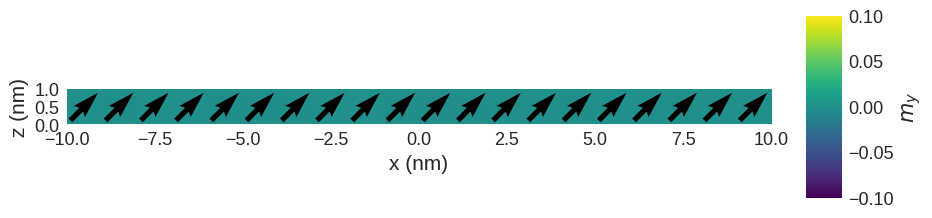
We can see that magnetisation is aligned with the sum of fields H1+H2. Finally, let us have a look at the table:
[9]:
system.table.data
[9]:
| max_mxHxm | E | delta_E | bracket_count | line_min_count | conjugate_cycle_count | cycle_count | cycle_sub_count | energy_calc_count | E_zeeman_zeeman1 | E_zeeman_zeeman2 | iteration | stage_iteration | stage | mx | my | mz | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.003228 | -2.843445e-20 | -7.222237e-35 | 27.0 | 5.0 | 21.0 | 23.0 | 2.0 | 33.0 | -1.421723e-20 | -1.421723e-20 | 30.0 | 30.0 | 0.0 | 0.707107 | 9.445158e-11 | 0.707107 |
We can see that energy terms are marked with the names we gave to energy terms when we defined the energy equation.


